The Open Source Movement in Digital Pathology AI
As COO of Tempus AI, I've had a front row seat on ways multimodal data and machine learning are transforming patient care. But few areas showcase the speed of transformation as dramatically as digital pathology. By converting pathology slides into gigapixel digital images, we've essentially created a new information source that machines can read, revealing patterns invisible to human eyes that can enable more precise, personalized cancer treatment decisions.
The AI models themselves are exciting, but what truly energizes me are the innovators contributing to the open-source community. Their collaborative approach is accelerating the entire field and bringing us closer to meaningful patient impact at unprecedented speed.
For healthcare AI leaders looking to leverage this open-source movement, I'll share a strategic framework at the end of this post focused on the intersection of open-source innovation and clinical deployment realities.
From Benchmarks to Breakthroughs
This journey began with watershed challenges like CAMELYON16, where researchers proved deep learning could detect cancer metastases with accuracy matching human pathologists. It wasn't just a technical achievement—it was proof that AI could truly "see" disease the way humans do.
That single moment changed everything. Subsequent challenges like CAMELYON17, TUPAC16, and PANDA built momentum by benchmarking mitosis detection, cancer grading, and biomarker prediction. Each breakthrough laid the groundwork for something bigger.
The Foundation Model Revolution
Today's digital pathology landscape looks radically different in the AI era. Foundation models, massive neural networks trained on millions of slide images, can now generalize across cancer types and diagnostic tasks. Instead of building custom models from scratch for each question, researchers start with broadly trained foundations and fine-tune them for specific diagnoses, prognoses, or even automated report generation.
What's remarkable is how much of this progress is being shared openly, and Paige helped pioneer this approach:
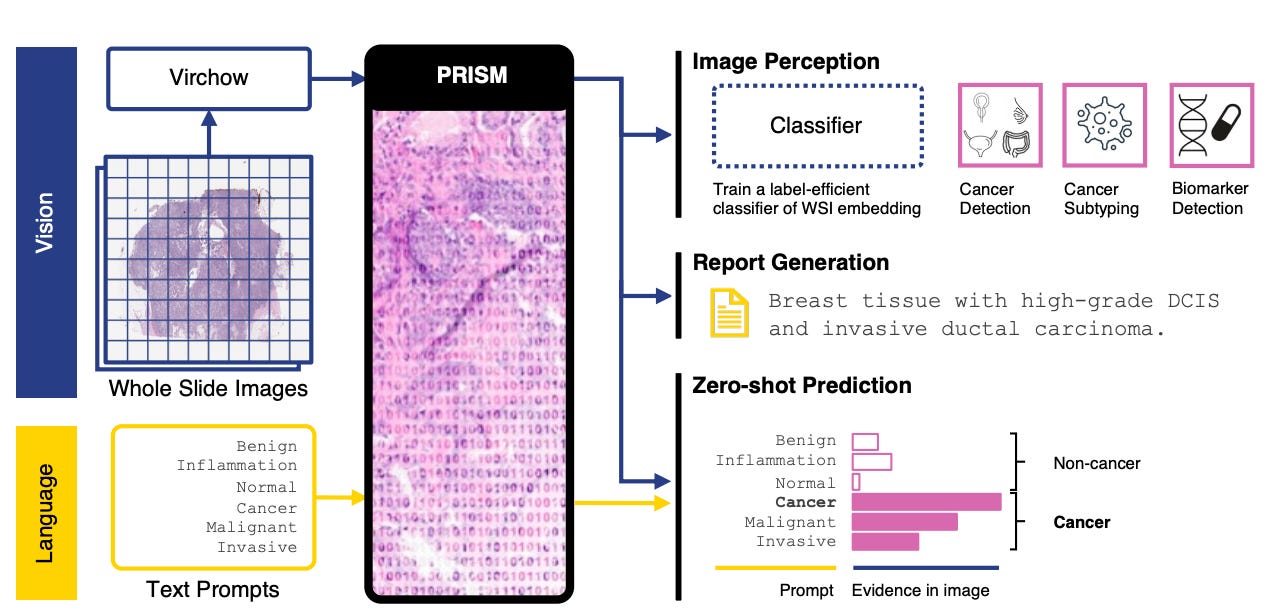
Paige's Virchow & PRISM: Virchow broke new ground as one of the first large pathology foundation models trained on over 1 million slides and openly released on Hugging Face. This was both unprecedented and a bold statement about the power of open collaboration to fuel digital pathology forward. PRISM extended this foundation to vision-language tasks, enabling draft report generation that bridges what AI sees with how pathologists communicate.
Microsoft & Providence's Prov-GigaPath (paper): Trained on 1.3 billion image tiles across 31 tissue types, this model outperformed predecessors on 25 of 26 benchmarks—including a 23% improvement in predicting EGFR gene mutations. The team released model weights openly, accelerating research worldwide.
Bioptimus H-Optimus-0 (press release): This 1.1-billion parameter model, trained on 500,000 slides from 4,000 clinics globally, set new performance benchmarks across diagnostic and prognostic tasks. Released for non-commercial use, it democratizes access to state-of-the-art pathology AI.
Academic Powerhouses: Models like UNI and CONCH from Mass General Brigham showcase how transformer-based architectures trained on 100,000+ slides achieve remarkable generalization across multiple cancer types.
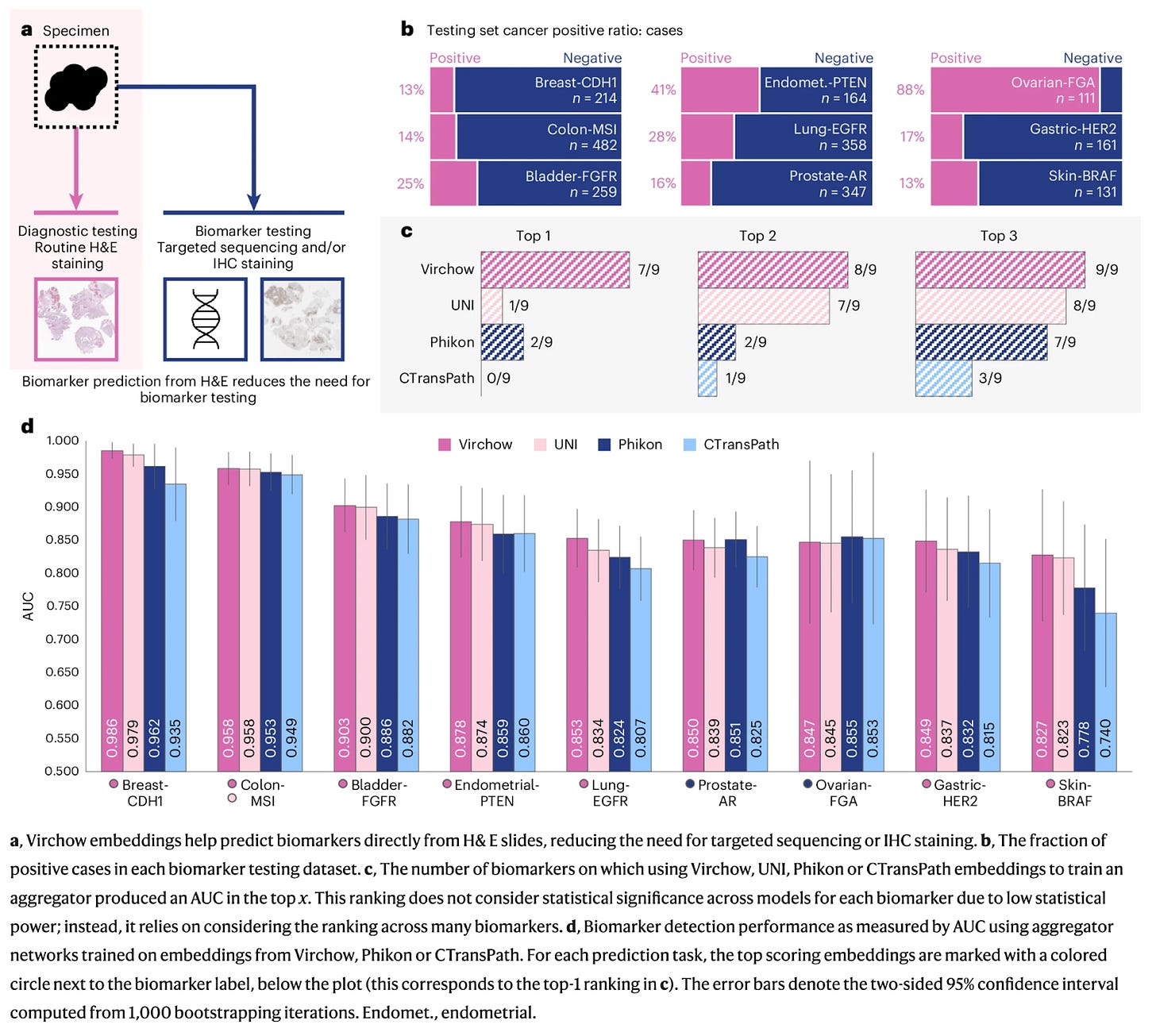
What the Benchmarks Reveal About the Future
A recent Nature paper benchmark compared leading public pathology foundation models across 22 clinical tasks from three health systems.
The results are striking:
Cancer detection is nearly a solved problem. Nearly all digital pathology foundation models achieved AUCs above 0.9 across detection tasks, with remarkably similar performance despite vastly different training approaches and dataset sizes. Whether you use a 22-million parameter model or a 1.1-billion parameter giant, the ability to spot cancer in tissue images has reached clinical-grade accuracy.
Biomarker and outcome prediction remains the next frontier. For tasks predicting treatment response, genetic mutations, and molecular biomarkers, performance varies dramatically between models. Unsurprisingly, models with greater representation of lung tissue, for example, performed far better on lung cancer biomarker tasks. The benchmark reveals that success in these challenging tasks depends not on raw computational power or model size, but on something far more nuanced—the composition and breadth of training data.
This shifts the conversation. The race isn’t about who can build the biggest model — it’s about who has the right data and the ability to integrate modalities beyond pathology slides.
The Network Effect in Action
This open-source momentum creates something powerful: a global ecosystem where academic labs, startups, healthcare providers, technology companies and pharmaceutical developers can build on each other's work rather than starting from zero.
Think about the implications: A researcher in Barcelona can now fine-tune Virchow for breast cancer detection using local patient data. A pharmaceutical company can leverage UNI to identify new biomarkers for drug trials. A pathologist in rural Montana gains access to the same diagnostic capabilities as major cancer centers.
The speed of innovation accelerates exponentially when the best models become the foundation rather than the finish line. Teams can shift resources from raw model development to clinical validation and workflow integration to ultimately have the biggest patient impact.
Building the Next Wave With The Tempus-Paige Advantage
Our recent acquisition of Paige isn't just about adding to our dataset or technology stack. It's about positioning ourselves at the forefront of the next wave of pathology AI to ensure every actionable biomarker is found to improve patient care.
Paige proved that boldness in open collaboration pays dividends. Virchow was the first million-slide foundation model openly released, establishing credibility and leadership. Paige also became the first to secure FDA clearance for a pathology AI product, which is critical regulatory experience that few others can claim.
But here's where the real strategic advantage emerges: While current benchmarks show that disease detection has reached clinical-grade accuracy across most models, they also reveal that biomarker prediction and treatment response require something fundamentally different. It requires the exact combination Tempus and Paige can deliver: models tuned with real-world multimodal datasets at unprecedented scale.
Performance on complex biomarker tasks correlates strongly with dataset composition and breadth, not just raw model size. Models trained on diverse, multimodal data consistently outperform those trained on larger but narrower image collections. Tempus brings real-world multimodal datasets spanning genomics, clinical outcomes, and imaging across thousands of institutions. Paige contributes 7 million clinically annotated pathology slides and the expertise to build technology that can work in a highly regulated healthcare market.
Together, we're uniquely positioned to build beyond the current foundation models benchmarks and establish the new standard for translating multimodal data into actionable decisions.
The Future We're Building Together
Digital pathology stands at an inflection point. Open-source foundation models are transforming isolated research projects into a connected global ecosystem.
But here's the key: building the most sophisticated models means nothing if they can't reach the patients who need them. Too often, promising AI research never makes the leap to actual real-world clinical impact. Models too often sit in research labs, trapped behind technical barriers, regulatory hurdles, or implementation challenges that make real-world deployment nearly impossible.
This is why we're not just focused on building better foundation models, we're building models that can actually be deployed in a highly regulated market like clinical care. Paige brings battle-tested regulatory expertise, having already navigated the complex FDA clearance process for pathology AI. They understand what it takes to move from proof-of-concept to patient impact. Tempus adds the clinical data infrastructure and healthcare system relationships that make large-scale deployment possible.
The future of precision medicine isn't just being built collaboratively, it requires a focused translation from an exciting model results into a deployable clinical-grade solution. We had to build the development infrastructure to translate our own promising AI applications into the clinical-grade AI diagnostics and now we’re excited to leverage those capabilities to enable others in the future.
For the 1% that have read this far, here’s my general framework for Healthcare AI Leaders interested in leveraging open-source models to impact patient care this is for you:
Framework for Healthcare AI Leaders
For those thinking about how to leverage open-source models for clinical impact, here’s my framework:
Build on Open Foundations – Don’t reinvent the wheel. Start with existing models to move as fast as possible.
Focus on Clinical Integration – Move resources to validation and workflow adoption.
Prioritize Multimodal Integration – Pathology + genomics + clinical = true precision medicine.
Scale Through Partnerships – Ecosystems accelerate innovation faster than solo efforts and partner with platforms for deployment into clinical workflows.
References:
Litjens et al., JAMA 2018 — CAMELYON16 challenge
Azizi et al., Nature 2024 — Prov-GigaPath
Vorontsov, E., Bozkurt, A., Casson, A. et al. Nature 2024 — Virchow & PRISM
Mass General Brigham Research — UNI and CONCH models
Bioptimus H-Optimus-0 — Foundation model release
If you found this post helpful, please share or subscribe to read more about the intersection of AI x Precision Medicine.


Thank you for the insightful post, Ryan! I appreciate your mention of H-optimus-0, which has topped the benchmark for diagnostics and biomarkers in the paper you mention. Just to clarify, H-optimus-0 is fully open source for both commercial and non-commercial applications (while its "big brother," H-optimus-1, trained on more diverse data and delivering increased performance, is available free of charge for non-commercial use).
I completely agree that the next frontier lies in training foundation models across different modalities and scales. This includes, e.g., jointly learning the molecular biology of tissues from single-cell and spatial transcriptomics data alongside tissue images. I'm excited to see the upcoming wave of innovations from companies like Tempus and Bioptimus, and as you highlight their impact on patients. Onwards!
Thanks Ryan. I am sure a lot went in to decide whether and how to acquire Paige over and above what you have described. I think you should have a ‘maker’ version of this ‘manager’ note and possibly a small community of makers will be grateful to learn from your pitch decks, IB notes, due diligence bcoz no one talks about it: how about you open source corporate development for pathology - a foundational model of getting FDA breakthrough designation or to raising series D in pathology based on so much you and Tempus have accomplished.